2. 中国农业科学院 农业资源与农业区划研究所,北京100081;
3. 盐城师范学院 海洋与生物工程学院,江苏 盐城 224002;
4. 南京农业大学 生命科学学院 农业部农业环境微生物工程重点开放实验室,江苏 南京 210095;
5. 江西省南康市教师进修学校,江西 赣州 341400
2. Institute of Agricultural Resources and Regional Planning, CAAS, Beijing 100081, China;
3. School of Marine and Biological Engineering, Yancheng Teachers University, Yancheng 224002, Jiangsu, China;
4. Key Laboratory of Microbiological Engineering of Agricultural Environment, Ministry of Agriculture, College of Life Science, Nanjing Agricultural University, Nanjing 210095, Jiangsu, China;
5. Nankang Teachers Training School, Ganzhou 341400, Jiangxi, China
化学农药是现代农业丰收的重要保障,其中除草剂是农药生产与使用的最大组成部分。虽然除草剂的长期、大量、广泛使用促进了粮食丰产,但也对农田及周边生态环境造成了不容忽视的危害,严重威胁人类的健康与生命安全,因此治理与修复除草剂的污染已引起了人们的广泛关注。生物修复(一般指微生物修复)因其具有高效、经济、无二次污染等优点在环境污染的治理中被广泛应用。自上世纪中叶开始,越来越多的农药降解菌被分离,对其降解代谢途径及关键降解基因的研究也取得了较大进展[1]。本文简述了目前全球广泛使用的8类除草剂的降解微生物资源、相关降解途径与降解基因的研究进展,并对除草剂污染的微生物修复研究与发展进行展望。
1 8类除草剂的微生物降解研究 1.1 有机磷类除草剂(以草甘膦为代表)草甘膦(glyphosate, GP)是目前世界上销售额最大的一种广谱、高效、非选择性、内吸型有机磷类除草剂,主要用于一年生和多年生以及单子叶和双子叶等40多个科的杂草的防治[2]。目前已分离到许多种属的草甘膦降解菌株,多数为细菌,来自不同的属,如假单胞菌属(Pseudomonas),苍白杆菌属(Ochrobacterum),节杆菌属(Arthrobacter),芽胞杆菌属(Bacillus),黄杆菌属(Flavobacterium),土壤杆菌属(Agrobacterium),根瘤菌属(Rhizobium),链霉菌属(Streptomyces),地芽胞杆菌属(Geobacillus),肠杆菌属(Enterobacter)等;少数真菌如曲霉属(Aspergillus),青霉菌属(Penicillum),帚霉属(Scopulariopsis),拟青霉属(Paecilomyces)和木霉属(Trichoderma)等[1]。其中人苍白杆菌(Ochrobactrum anthropi)GPK3和无色杆菌(Achromobacter sp.)Kg16[3]分离自受草甘膦污染的土壤,能够利用草甘膦作为磷源生长;枯草芽胞杆菌Bs-15(Bacillus subtilis)[2]分离自胡椒根际土壤,在高浓度草甘膦的培养基中生长良好,最高耐受浓度达40 000 mg/L,在未灭菌土壤中96 h内对10 000 mg/L草甘膦的降解效率达71.57%;土壤放射杆菌(Agrobacterium radiobacter)SW9[4]来源于草甘膦废水处理系统的活性污泥,能够利用草甘膦为唯一碳源和能源;链霉菌StC[5]分离自污水处理厂活性污泥,能够利用草甘膦作为唯一碳源、氮源和磷源生长。
草甘膦在微生物作用下降解的起始途径共有3种:第一种是通过草甘膦氧化还原酶(Gox)的氧化作用催化草甘膦的C—N键断裂生成氨基甲基磷酸和乙醛酸,从而使其降解[6, 7];第二种是来自惠特莫尔氏杆菌(Pseudomonas pseudomallei)的基因glpA/glpB编码表达的裂解酶特异催化草甘膦的C—P键断裂生成肌氨酸,随后在肌氨酸氧化酶作用下进一步转化为甘氨酸[5];第三种途径是由Castle等[8]发现的一种抗草甘膦基因草甘膦N-乙酰转移酶GAT,在该酶作用下催化草甘膦转化为脱毒的乙酰化草甘膦。
目前国内外已经商业化种植的转基因抗草甘膦作物均采用一种来自于根癌农杆菌(Agrobacterium tumefaciens)CP4的具有天然草甘膦抗性的烯醇丙酮酸磷酸莽草酸(EPSP)合酶(CP4-EPSPS)[9],该酶的草甘膦耐受能力十分突出,其Ki/Km值超过了200,从目前资料看,还没有发现其他抗性能力接近CP4-EPSPS的酶。另外美国孟山都公司研发的含有CP4-EPSPS和GOX(专利US5776760)[10]的转基因GT73油菜已经进入商业化应用阶段。先锋公司正在致力于改造优化gat基因以期尽快用于转基因作物的构建。
1.2 磺酰脲类除草剂磺酰脲类除草剂是世界上使用量最大的第二大类除草剂,仅次于草甘膦,目前已有30多个商品化产品。常见的有甲磺隆、苯磺隆、苄嘧磺隆、氯嘧磺隆、烟嘧磺隆、胺苯磺隆等[11],脲桥结构是磺酰脲类除草剂的典型结构(图 1)。
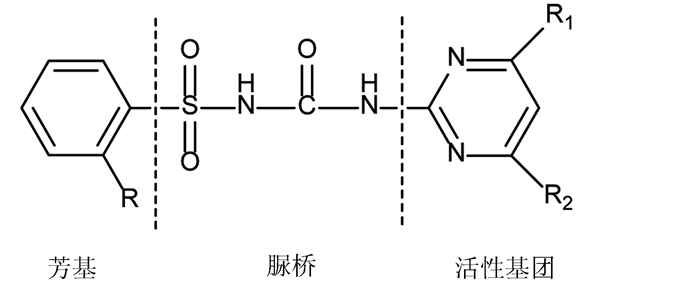
|
图 1 磺酰脲类除草剂的典型结构通式 Figure 1 Representative structure formula of sulfonylurea herbicides |
目前已分离到多株磺酰脲类除草剂降解菌株,其中大多数降解菌以断裂脲桥C—N键的方式为主,如氯嘧磺隆降解菌克雷伯氏菌(Klebsiella jilinsis)2N3分离自工业废水处理的活性污泥,对起始浓度为100 mg/L的氯嘧磺隆在12 h内的降解率为92.5%,并且该菌还能同时降解烟嘧磺、甲磺隆、苯磺隆、胺苯磺隆和砜嘧磺隆[12];甲磺隆降解菌株甲基营养菌株(Methylopila sp.)S113分离自被甲磺隆污染的土壤,其在72 h内对50 mg/L的甲磺隆的降解率高达97%,同时也能够降解苄嘧磺隆,噻吩磺隆和胺苯磺隆[13]。
目前已报道的磺酰脲类除草剂降解基因较少。其中从浅灰色链霉菌(Streptomyces griseolus) ATCC 11796的基因组中克隆到一个能催化多种磺酰脲类除草剂羟基化反应的细胞色素P450Su1基因,该基因编码表达一个由细胞色素P450氧化酶、铁氧还蛋白和NAD(P)H铁氧还蛋白还原酶组成的三组分氧化还原体系,将P450Su1基因转入烟草发现转基因烟草表达的P450su1催化磺酰脲类除草剂R7402的N-脱烷基反应后形成对植物毒性更强的代谢产物[14, 15]。另外从甲基营养菌株S113的基因组克隆到一个酯酶基因sulE(又名tsmE,专利号ZL20111020744.2)。SulE降解谱广,可将噻吩磺隆、甲磺隆、氯嘧磺隆、苄嘧磺隆、胺苯磺隆等磺酰脲除草剂的酯键水解,生成相应的无除草活性酸产物。SulE分子量较小,具有较宽的pH和温度耐受范围,且其催化活性不需要任何辅因子。将sulE导入到酵母菌(Saccharomyces cerevisiae)BY4741可显著提高该菌对噻吩磺隆和甲磺隆的耐受性,表明SulE是一个理想的抗除草剂转基因工程的除剂脱毒酯酶。目前sulE能否提高植物对磺酰脲类除草剂耐受性的工作还未见报道。
1.3 氯乙酰胺类除草剂氯乙酰胺类除草剂是一类分子结构有氯乙酰胺基团的高效除草剂,使用量居世界第三位,主要品种包括乙草胺、丁草胺、甲草胺、异丙甲草胺和异丙草胺。国内外已分离筛选到多株该类除草剂降解菌株,如分离自水稻田土壤的副球菌(Paracoccus sp.) FLY-8能够同时降解并利用6种氯乙酰胺类除草剂作为碳源生长,并因除草剂分子结构中侧链的差异而具有不同的降解速率,依次为甲草胺>乙草胺>丙草胺>丁草胺>丙草胺>异丙甲草胺[17];乙草胺降解菌株鞘氨醇单胞菌(Sphingomonas sp.)DC-6[18]分离自农药厂污水处理的活性污泥,能够完全矿化甲草胺,乙草胺和丁草胺,48 h内对甲草胺,乙草胺和丁草胺的降解率分别为98.6%,93.7%和76.7%,包括红球菌(Rhodococcus sp.)T3-1,戴尔福特菌(Delftia sp.) T3-6和鞘脂菌(Sphingobium sp.)MEA3-1[19]的菌群分离自受乙草胺污染土壤,能够通过共代谢的方式联合矿化甲草胺,乙草胺和丁草胺;鞘脂菌DC-2[20]与鞘脂菌(Sphingobium baderi) DE-13分离自活性污泥,能够联合矿化乙草胺,其中菌株DC-2只负责将乙草胺降解至2-甲基-6-乙基苯胺(MEA),随后菌株DE-13负责MEA的完全降解。
氯乙酰胺类除草剂的微生物降解途径主要有两种(图 2):谷胱甘肽-共轭脱氯途径[21]和N-脱烷基氧化途径[18, 19]。第一种谷胱甘肽-共轭脱氯途径以甲草胺为例,其在谷胱甘肽转移酶的催化作用下,共轭脱氯形成甲草胺-半胱氨酸结合物,随后该结合物在羧肽酶、γ-谷酰胺转肽酶和半胱氨酸-β-裂解酶的共同作用下,硫原子被氧化为磺酸基团,形成中间产物甲草胺-甲磺酸(甲草胺-ESA)[21]。第二种N-脱烷基氧化途径以乙草胺为例,其在N-脱烷基酶CndABC(来自鞘氨醇单胞菌DC-6)[22]/EthBAD(来自红球菌T3-1)[19]的催化作用下脱去乙氧甲基基团,生成2-甲基-6-乙基苯基乙酰胺(CMEPA),CMEPA在酰胺水解酶CmeH(来自鞘脂菌DC-2)[20]/ DamH(来自戴尔福特菌T3-6)[23]的作用下转化为MEA,随后MEA在单加氧酶MeaXY[24]的羟基化作用下被氧化为MEA-OH。在鞘脂菌MEA3-1中,MEA-OH的自发氧化形成2-甲基-6-乙基对苯二酚(MEHQ),然后MEHQ在MeaAB的氧化作用下转化为3-OH-MEHQ,最终被降解至CO2和水[25]。
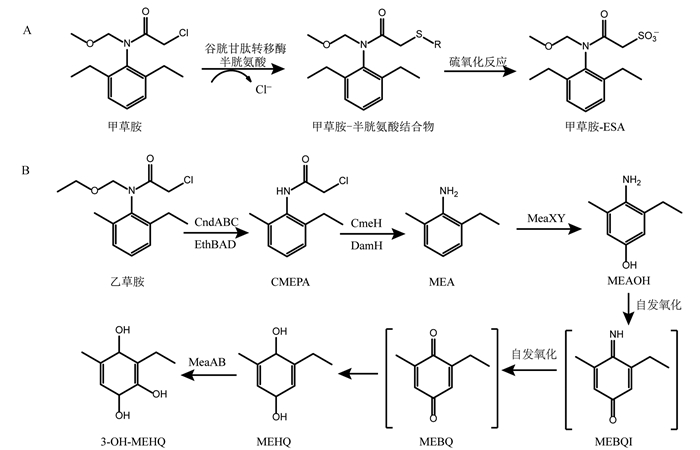
|
图 2 氯乙酰胺类除草剂的降解途径及相关的降解酶[15, 16, 18] Figure 2 Degradation pathways of chloroacetyl herbicides and related degradation enzymes[15, 16, 18] 注:A, 甲草胺-谷胱甘肽共轭脱氯途径;B, 乙草胺N-脱烷基氧化途径 Note:A, conjugate dechlorination of alachlor-glutathione; B, acetochlor degradation by N-dealkylation pathway |
以莠去津为代表的三嗪类除草剂是一类传统的除草剂,主要用于玉米、甘蔗和草皮等作物田地防除阔叶与禾本科杂草[26]。假单胞菌ADP是首个报道的莠去津降解菌株,能在24 h内完全降解100 mg/L的莠去津,能利用莠去津为唯一氮源生长,并且对高浓度的莠去津(浓度大于1 000 mg/L)有较强的耐受性[27]。另外,有报道一个8成员混合菌群共代谢莠去津,该菌群包括根癌农杆菌,新月柄杆菌(Caulobacter crescentus),恶臭假单胞菌(Pseudomonas putida),鞘氨醇单胞菌(Sphingomonas yaniokuyae),诺卡氏菌(Nocardia sp.),根瘤菌,栖稻黄单胞菌(Flavobacterium oryzihabitans)和产碱菌(Variovorax paradoxus)[28]。许多均三嗪类降解菌株能够同时降解多种此类除草剂,如类诺卡氏菌(Nocardioides sp.)DN36分离自施用西草净的水稻田土壤,它能够完全降解西草净、莠去津、西玛津和扑灭津,也可以转化莠灭净,异戊净等,是首个报道的能够单独开环矿化多种均三嗪类除草剂的降解菌株[29]。
莠去津的降解起始途径主要有两种(图 3),第一种途径是其在脱氯酶AtzA(主要在革兰氏阴性菌中)或TrzN(主要为革兰氏阳性菌)[30]的水解作用下脱去氯原子生成羟基化莠去津,然后在水解酶AtzB和AtzC作用下逐步被转化为三聚氰酸;第二种降解起始途径是莠去津在未知N-脱烷基酶的作用下脱去乙基和异丙基生成2-氯-4-羟基-6-氨基-1, 3, 5-三嗪,最终也被转化为三聚氰酸。莠去津的下游产物三聚氰酸在酰胺水解酶AtzD的催化作用下开环生成缩二脲,缩二脲在水解酶AtzE的作用下转化为脲基甲酸,最终在水解酶AtzF的作用下降解为CO2和氨气[31]。
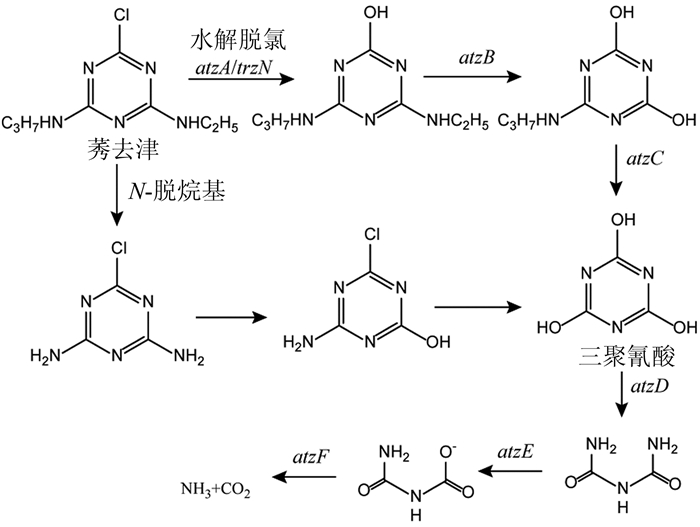
|
图 3 莠去津的降解途径及降解基因[26, 27] Figure 3 Catabolic pathways of atrazine and related degradation genes[26, 27] |
目前,针对均三嗪类除草剂的污染采用原位投加降解菌体的生物修复技术已有很多研究[32],如对莠去津降解基因atzA转基因作物如苜蓿、烟草和拟南芥的植物-微生物修复正引起越来越多的关注[33]。
1.5 芳氧基苯氧基丙酸酯类除草剂芳氧基苯氧基丙酸酯类(AOPP)除草剂通过抑制杂草中乙酰辅酶A羧化酶的活性高效防除禾本科杂草,常见的商品化产品有禾草灵、精恶唑禾草灵和氰氟草酯等。此类除草剂微生物降解的第一步反应均是在酯酶的催化作用下发生酯键断裂,生成相应的酸式除草活性物质(图 4)。目前已克隆到4个不同的编码水解酯酶的基因,分别是:feH,来自红球菌T1[34]和赤红球菌(Rhodococcus ruber)JPL-2[35];afeH, 来自不动杆菌(Acinetobacter sp.)DL-2[36];chbH, 来自假单胞菌QDZ-1[37];fpbH, 来自水微菌(Aquamicrobium sp.) FPB-1[38]。这些酯酶一般能够同时水解多种类型的AOPP除草剂,但这些除草剂中的醇烷基侧链的长度影响酯酶的水解活性,如由fpbH编码的酯酶FpbH对AOPP类除草剂水解活性大小为精吡氟氯禾灵>禾草灵>精恶唑禾草灵>精喹禾灵>精吡氟禾草灵>氰氟草酯[38]。
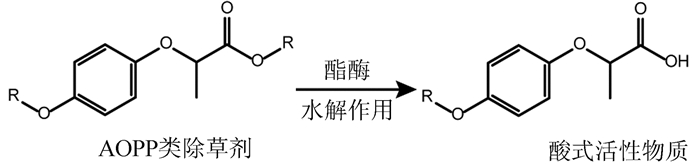
|
图 4 AOPP类除草剂结构通式及其水解降解途径 Figure 4 Basic structure and hydrolytic degradation pathway of AOPP herbicides |
苯氧乙酸类除草剂是一种激素类除草剂,广泛用于小麦、玉米和水稻田阔叶杂草的防除,代表产品为2, 4-D。目前已报道的2, 4-D降解微生物包括真菌、细菌和放线菌,其中细菌占据主导地位[39]。微生物降解2, 4-D主要通过两条代谢途径:①在α-酮戊二酸依赖型双加氧酶TfdA(来自富养产碱菌JMP134)[40]或CadABC(来自短根瘤菌HW13)[39]的作用下,催化2, 4-D脱去乙酸基团生成毒性比2, 4-D低100倍的2, 4-二氯苯酚,随后2, 4-二氯苯酚在羟化酶TfdB的催化下被转化为3, 5-二氯邻苯二酚,然后依次在TfdCDEF的作用下开环最终被降解为β-酮己二酸,进入三羧酸循环;②在圆褐固氮菌(Azotobacter chroococcum)中,2, 4-D在未知脱氯酶的作用下首先脱去一个氯原子生成对氯苯氧乙酸,随后氧化脱去乙酸基团生成对氯苯酚,进一步羟基化生成4-氯邻苯二酚,然后开环被降解为β-酮己二酸,最终进入三羧酸循环[41]。
陶氏益农公司将来源于土壤细菌Sphingobium herbicidovorans Mh的编码芳氧基链烷酸酯双加氧酶基因aad-1应用于多种抗2, 4-D作物的研发[42, 43],其中耐2, 4-D玉米DAS-40278-9[44]已获得美国农业部(USDA)商业化种植和食品与药物管理局(FDA)食用和使用批准,并在加拿大、澳大利亚和巴西等国家进行商业化推广和种植,目前该转基因玉米正在申请我国国内用作加工原料的安全证书(http://www.moa.gov.cn/ztzl/zjyqwgz/spxx/201307/P020170802513991980165.pdf)。陈志贤等[45]将2, 4-D单氧化酶tfdA导入晋棉7号、冀合321等棉花品种,其后代田间抗药性鉴定结果表明转基因系对2, 4-D的耐受能力远超过大田使用浓度。
1.7 二硝基苯胺类除草剂(以二甲戊乐灵为代表)二硝基苯胺除草剂广泛用于棉花、大豆和玉米田一年生禾本科杂草的防除,最常用的包括二甲戊乐灵、仲丁灵、氟乐灵和氨磺乐灵。研究表明在好氧条件下微生物主要通过硝基还原[46]、N-脱烷基氧化[47]和环化作用[48]降解二甲戊灵和氟乐灵。枯草芽胞杆菌Y3分离自活性污泥,在2.5 d内对100 mg/L的二甲戊灵的降解率为99.5%,并能够以二甲戊灵为唯一碳源生长[46]。菌株Y3通过硝基还原酶PNR将二甲戊灵还原至6-氨基二甲戊灵,然后在相关酶的作用下进一步被硝基还原羧化、环化,形成新的中间产物(图 5)[49]。
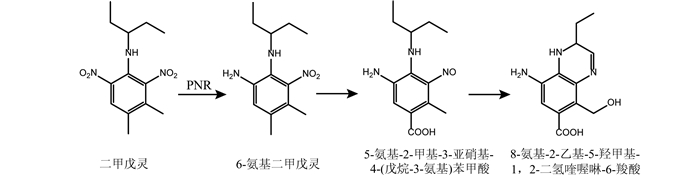
|
图 5 Bacillus subtilis Y3的二甲戊灵降解途径[49] Figure 5 Degradation pathway of pendimethalin in Bacillus subtilis Y3[49] |
硫代氨基甲酸酯类除草剂是一类内吸传导型苗前除草剂,主要用于水稻田防除一年生禾本科杂草,特别是对稗草的防治具有特效。使用较为广泛的产品有杀草丹、禾草敌和野麦畏等。微生物是杀草丹在环境中降解的主要因素,目前已报道的杀草丹降解菌株有食酸菌(Acidovorax sp.)T1[50],棒状杆菌(Corynebacterium sp.)[51]和黑曲霉(Aspergillus niger)CRN[52]。食酸菌T1分离自农药厂活性污泥,在36 h内对约100 mg/L杀草丹的降解率达到95.4%,且能够利用杀草丹作为碳源,氮源和硫源生长。在菌株T1中,杀草丹在双组份的FMN依赖型单加氧酶TmoAB的作用下,断C—S键氧化至对氯苯甲醛和N, N-二乙基硫代氨基甲酸,随后对氯苯甲醛在脱氢酶TmoC的作用下被氧化为对氯苯甲酸,对氯苯甲酸依次在脱氯酶FcbABC,单加氧酶PobA的催化作用下水解脱氯、羟基化反应至原儿茶酸,经原儿茶酸4, 5-双加氧酶LigAB开环途径依次被降解至草酰乙酸和丙酮酸并进入三羧酸循环[50](图 6)。
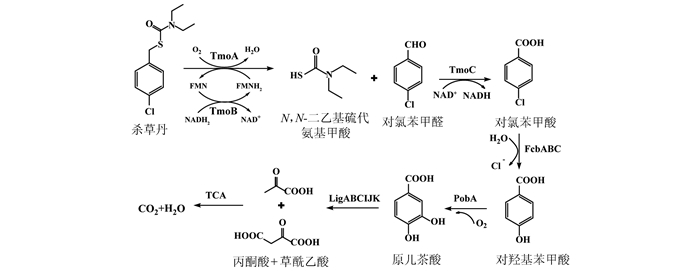
|
图 6 菌株Acidovorax sp.T1降解杀草丹的代谢途径及相关降解酶[49] Figure 6 Degradation pathway of thiobencarb and related enzymes in Acidovorax sp.T1[49] |
禾草敌的微生物降解研究较为清楚的是一个5成员的微生物菌群DC的共代谢途径,由绿针假单胞菌(Pseudomonas chlororaphis)ON1,嗜麦芽寡养单胞菌(Stenotrophomonas maltophilia)ON2,硝基还原假单胞菌(Pseudomonas nitroreducens)ON3,喜珍品杆菌(Gulosibacter molinativorax)ON4和木糖氧化无色杆菌(Achromobacter xylosoxidans)ON5[53]组成。其中菌株ON4通过水解酶MolA[54]断硫酯键将禾草敌降解为乙硫醇和氮杂环庚烷-1-羧酸叔丁酯,随后菌株ON1、ON3和ON4共同负责氮杂环庚烷-1-羧酸叔丁酯的降解,乙硫醇的降解则由菌株ON1和ON2完成[55]。
2 展望微生物数量与种类的繁多及其代谢类型的多样性使其在农药生物降解方面占据绝对优势,分离和筛选除草剂高效降解菌株为受污染土壤、水环境等的生物修复提供了有力保障。本文简要介绍了使用较为广泛的除草剂中代表产品的降解微生物及相关降解途径和降解基因。这些已分离的降解微生物组成多样,其中细菌占据了大部分,其次是真菌和放线菌。已经报道的除草剂降解途径多是基于降解中间代谢产物进行的推测,绝大部分的降解基因没有被克隆,或者仅克隆到参与部分降解的基因,缺乏对菌株降解特性和降解机制的全面和深入了解,从而限制了降解菌剂、降解基因和降解酶资源的开发和应用。
近年来,通过引入微生物除草剂降解基因,构建转基因作物来提高农作物的除草剂抗性,或者利用转基因植物进行原位修复正成为除草剂污染治理的另一个重要发展方向。但是转基因作物的构建和应用还存在很大的局限性,微生物降解过程需要一系列基因和酶的作用与调控,而当前条件下多是转单个基因至作物体内,只能将除草剂部分转化至其下游代谢产物,还不能完全解除除草剂毒害。因此构建具备完善降解体系的转基因作物将面临着挑战,这也使得进一步探究微生物的完整降解途径及克隆整套降解基因的研究更加迫切。
| [1] |
Huang X, He J, Yan X, et al. Microbial catabolism of chemical herbicides: microbial resources, metabolic pathways and catabolic genes[J]. Pestic Biochem Phys, 2016, 143: 272-297. |
| [2] |
Yu X M, Yu T, Yin G H, et al. Glyphosate biodegradation and potential soil bioremediation by Bacillus subtilis strain Bs-15[J]. Genet Mol Res, 2015, 14(4): 14 717-14 730. DOI:10.4238/2015.November.18.37 |
| [3] |
Ermakova I T, Shushkova T V, Sviridov A V, et al. Organophosphonates utilization by soil strains of Ochrobactrum anthropi and Achromobacter sp.[J]. Arch Microbiol, 2017, 199(5): 665-675. DOI:10.1007/s00203-017-1343-8 |
| [4] |
Mcauliffe K S, Hallas L E, Kulpa C F. Glyphosate degradation by Agrobacterium radiobacter isolated from activated sludge[J]. J Ind Microbiol Biotechnol, 1990, 6(3): 219-221. |
| [5] |
Obojska A, Lejczak B, Kubrak M. Degradation of phosphonates by streptomycete isolates[J]. Appl Microbiol Biotechnol, 1999, 51(6): 872-876. DOI:10.1007/s002530051476 |
| [6] |
Gard J K, Feng P C, Hutton W C. Nuclear magnetic resonance timecourse studies of glyphosate metabolism by microbial soil isolates[J]. Xenobiotica, 1997, 27(7): 633-644. DOI:10.1080/004982597240235 |
| [7] |
Plinesrnic W. Physiological mechanisms of glyphosate resistance[J]. Weed Technol, 2006, 20(2): 290-300. DOI:10.1614/WT-04-131R.1 |
| [8] |
Castle L A, Siehl D L, Gorton R, et al. Discovery and directed evolution of a glyphosate tolerance gene[J]. Science, 2004, 304(5674): 1 151-1 154. DOI:10.1126/science.1096770 |
| [9] |
Barry G F, Kishore G M, Padgette S R, et al. Glyphosate-tolerant 5-enolpyruvylshikimate-3-phosphate synthases: US, US 6248876 B1[P]. 2001.
|
| [10] |
Barry G F, Kishore G M. Glyphosate tolerant plants: EP, US RE38825 E1 [P]. 2005.
|
| [11] |
Zhang M H. The development situation, market and trend of sulfonylurea herbicides[J]. Agrochemicals, 2010, 49(4): 235-240, 245. 张敏恒. 磺酰脲类除草剂的发展现状、市场与未来趋势[J]. 农药, 2010, 49(4): 235-240, 245. |
| [12] |
Zhang H, Zhang X, Mu W, et al. Biodegradation of chlorimuron-ethyl by the bacterium Klebsiella jilinsis 2N3[J]. J mental Sci Heal B, 2010, 45(6): 501-507. DOI:10.1080/03601234.2010.493473 |
| [13] |
Huang X, He J, Sun J, et al. Isolation and characterization of a metsulfuron-methyl degrading bacterium Methylopila sp. S113[J]. Int Biodeterior Biodegr, 2007, 60(3): 152-158. DOI:10.1016/j.ibiod.2007.02.005 |
| [14] |
O'keefe D P, Romesser J A, Leto K J. Identification of constitutive and herbicide inducible cytochromes P-450 in Streptomyces griseolus[J]. Arch Microbiol, 1988, 149(5): 406-412. DOI:10.1007/BF00425579 |
| [15] |
Omer C A, Lenstra R, Litle P J, et al. Genes for two herbicide-inducible cytochromes P-450 from Streptomyces griseolus[J]. J Bacteriol, 1990, 172(6): 3 335-3 345. DOI:10.1128/jb.172.6.3335-3345.1990 |
| [16] |
Hang B J, Hong Q, Xie X T, et al. SulE, a sulfonylurea herbicide de-esterification esterase from Hansschlegelia zhihuaiae S113[J]. Appl Environ Microbiol, 2012, 78(6): 1 962-1 968. DOI:10.1128/AEM.07440-11 |
| [17] |
Zhang J, Zheng J W, Liang B, et al. Biodegradation of chloroacetamide herbicides by Paracoccus sp. FLY-8 in vitro[J]. J Agr Food Chem, 2011, 59(9): 4614-4621. DOI:10.1021/jf104695g |
| [18] |
Chen Q, Yao L, Wang C H, et al. Isolation and characterization of acetochlor-degrading strain Sphingomonas sp. DC-6 and preliminary studies on its metabolic pathway[J]. J Agr Sci Tech, 2013, 15(5): 67-74. |
| [19] |
Wang F, Zhou J, Li Z, et al. Involvement of the cytochrome P450 system EthBAD in the N-deethoxymethylation of acetochlor by Rhodococcus sp.strain T3-1[J]. Appl Environ Microbiol, 2015, 81(6): 2182-2188. DOI:10.1128/AEM.03764-14 |
| [20] |
Li Y, Chen Q, Wang C H, et al. Degradation of acetochlor by consortium of two bacterial strains and cloning of a novel amidase gene involved in acetochlor-degrading pathway[J]. Bioresour Technol, 2013, 148(11): 628-631. |
| [21] |
Stamper D M, Tuovinen O H. Biodegradation of the acetanilide herbicides alachlor, metolachlor, and propachlor[J]. Crit Rev Microbiol, 1998, 24(1): 1-22. DOI:10.1080/10408419891294163 |
| [22] |
Chen Q, Wang C H, Deng S K, et al. Novel three-component Rieske non-heme iron oxygenase system catalyzing the N-dealkylation of chloroacetanilide herbicides in sphingomonads DC-6 and DC-2[J]. Appl Environ Microbiol, 2014, 80(16): 5 078-5085. DOI:10.1128/AEM.00659-14 |
| [23] |
Wang F, Hou Y, Zhou J, et al. Purification of an amide hydrolase DamH from Delftia sp. T3-6 and its gene cloning, expression, and biochemical characterization[J]. Appl Microbiol Biotechnol, 2014, 98(17): 7491-7499. DOI:10.1007/s00253-014-5710-y |
| [24] |
Cheng M, Qiang M, Yang Y, et al. The two-component monooxygenase MeaXY initiates the downstream pathway of chloroacetanilide herbicide catabolism in Sphingomonads[J]. Appl Environ Microbiol, 2017, 83(7): e03 241-03 216. |
| [25] |
Dong W L, Chen Q Z, Hou Y, et al. Metabolic pathway involved in 2-methyl-6-ethylaniline degradation by Sphingobium sp.strain MEA3-1 and cloning of the novel flavin-dependent monooxygenase system meaBA[J]. Appl Environ Microbiol, 2015, 81(24): 8254-8264. DOI:10.1128/AEM.01883-15 |
| [26] |
Liu T, Cao P, Geng J, et al. Determination of triazine herbicides in milk by cloud point extraction and high-performance liquid chromatography[J]. Food Chem, 2014, 142(3): 358. |
| [27] |
de Souza M L, Wackett L P, Sadowsky M J. The atzABC genes encoding atrazine catabolism are located on a self-transmissible plasmid in Pseudomonas sp. strain ADP[J]. Appl Environ Microbiol, 1998, 64(6): 2323. |
| [28] |
Smith D, Alvey S, Crowley D E. Cooperative catabolic pathways within an atrazine-degrading enrichment culture isolated from soil[J]. FEMS Microbiol Ecol, 2005, 53(2): 265-273. DOI:10.1016/j.femsec.2004.12.011 |
| [29] |
Satsuma K. Mineralization of s-triazine herbicides by a newly isolated Nocardioides species strain DN36[J]. Appl Microbiol Biotechnol, 2010, 86(5): 1 585-1 592. DOI:10.1007/s00253-010-2460-3 |
| [30] |
Omotayo A E, Ilori M O, Amund O O, et al. Establishment and characterization of atrazine degrading cultures from Nigerian agricultural soil using traditional and Bio-Sep bead enrichment techniques[J]. Appl soil ecol, 2011, 48(1): 63-70. DOI:10.1016/j.apsoil.2011.01.006 |
| [31] |
Hernández M, Jia Z, Conrad R, et al. Simazine application inhibits nitrification and changes the ammonia-oxidizing bacterial communities in a fertilized agricultural soil[J]. FEMS Microbiol Ecol, 2011, 78(3): 511-519. DOI:10.1111/fem.2011.78.issue-3 |
| [32] |
Morgante V, Lópezlópez A, Flores C, et al. Bioaugmentation with Pseudomonas sp. strain MHP41 promotes simazine attenuation and bacterial community changes in agricultural soils[J]. FEMS Microbiol Ecol, 2010, 72(1): 152-152. DOI:10.1111/fem.2010.72.issue-1 |
| [33] |
Wang L, Samac D A, Shapir N, et al. Biodegradation of atrazine in transgenic plants expressing a modified bacterial atrazine chlorohydrolase (atzA) gene[J]. Plant Biotechnol J, 2010, 3(5): 475-486. |
| [34] |
Hou Y, Tao J, Shen W, et al. Isolation of the fenoxaprop-ethyl (FE)-degrading bacterium Rhodococcus sp. T1, and cloning of FE hydrolase gene feh[J]. Fems Microbiology Letters, 2011, 323(2): 196-203. DOI:10.1111/fml.2011.323.issue-2 |
| [35] |
Liu H M, Lou X, Ge Z J, et al. Isolation of an aryloxyphenoxy propanoate (AOPP) herbicide-degrading strain Rhodococcus ruber JPL-2 and the cloning of a novelcarboxylesterase gene (feh)[J]. Braz J Microbiol, 2015, 46(2): 425-432. DOI:10.1590/S1517-838246220140208 |
| [36] |
Dong W, Jiang S, Shi K, et al. Biodegradation of fenoxaprop-P-ethyl (FE) by Acinetobacter sp. strain DL-2 and cloning of FE hydrolase gene afeH[J]. Bioresour Technol, 2015, 186: 114-121. DOI:10.1016/j.biortech.2015.03.039 |
| [37] |
Nie Z J, Hang B J, Cai S, et al. Degradation of cyhalofop-butyl (CyB) by Pseudomonas azotoformans strain QDZ-1 and cloning of a novel gene encoding CyB-hydrolyzing esterase[J]. J Agric Food Chem, 2011, 59(11): 6 040-6 046. DOI:10.1021/jf200397t |
| [38] |
Wang C H, Qiu J G, Yang Y J, et al. Identification and characterization of a novel carboxylesterase (FpbH) that hydrolyzes aryloxyphenoxypropionate herbicides[J]. Biotechnol Lett, 2017, 39(4): 553-560. DOI:10.1007/s10529-016-2276-z |
| [39] |
Kitagawa W, Takami S, Miyauchi K, et al. Novel 2, 4-dichlorophenoxyacetic acid degradation genes from oligotrophic Bradyrhizobium sp. strain HW13 isolated from a pristine environment[J]. J Bacteriol, 2002, 184(2): 509-518. DOI:10.1128/JB.184.2.509-518.2002 |
| [40] |
Don R H, Weightman A J, Knackmuss H J, et al. Transposon mutagenesis and cloning analysis of the pathways for degradation of 2, 4-dichlorophenoxyacetic acid and 3-chlorobenzoate in Alcaligenes eutrophus JMP134(pJP4)[J]. J Bacteriol, 1985, 161(1): 85-90. |
| [41] |
Balajee S, Mahadevan A. Dissimilation of 2, 4-dichlorophenoxyacetic acid by Azotobacter chroococcum[J]. Xenobiotica, 1990, 20(6): 607-617. DOI:10.3109/00498259009046876 |
| [42] |
Wright T R, Shan G, Walsh T A, et al. Robust crop resistance to broadleaf and grass herbicides provided by aryloxyalkanoate dioxygenase transgenes[J]. Proc Natl Acad Sci U S A, 2010, 107(47): 20240-20245. DOI:10.1073/pnas.1013154107 |
| [43] |
Skelton J J, Simpson D M, Peterson M A, et al. Comparative analysis of 2, 4-D uptake, translocation, and metabolism in non-AAD-1 transformed and 2, 4-D-resistant corn[J]. Weed Science, 2017, 1-12. |
| [44] |
Stagg N J, Thomas J, Herman R A, et al. Acute and 28-day repeated dose toxicology studies in mice with aryloxyalkanoate dioxygenase (AAD-1) protein expressed in 2, 4-D tolerant DAS-40278-9 maize[J]. Regul Toxicol Pharmacol, 2012, 62(2): 363-370. DOI:10.1016/j.yrtph.2011.10.018 |
| [45] |
Chen Z X, Llewellyn D J, Fan Y L, et al. Heritable transgenic 2, 4-D-resistant cotton obtained by transformation of tfdA mediated by Agrobacterium tumefaciens[J]. Scientia Agricutura Sinica, 1994, 27(2): 31-37. 陈志贤, 范云六, 李淑君, 等. 利用农杆菌介导法转移tfdA基因获得可遗传的抗2, 4-D棉株[J]. 中国农业科学, 1994, 27(2): 31-37. |
| [46] |
Ni H Y, Yao L, Li N, et al. Biodegradation of pendimethalin by Bacillus subtilis Y3[J]. J Environ Sci (China), 2016, 41(3): 121-127. |
| [47] |
More V S, Tallur P N, Niyonzima F N, et al. Enhanced degradation of pendimethalin by immobilized cells of Bacillus lehensis XJU[J]. Biotech, 2015, 5(6): 1-8. |
| [48] |
Sharef I B, Abdelbagi A O, Elsheikh E A E, et al. Biodegradation of pendimethalin by three strains of bacteria isolated from pesticide-polluted soils[J]. U of K J Agric Sci, 2015(2): 233-252. |
| [49] |
Ni H Y, Wang F, Li N, et al. Pendimethalin nitroreductase is responsible for the initial pendimethalin degradation step in Bacillus subtilis Y3[J]. Appl Environ Microbiol, 2016, 82(24): 7 052-7062. DOI:10.1128/AEM.01771-16 |
| [50] |
Chu C W, Liu B, Li N, et al. A novel aerobic degradation pathway of thiobencarb is initiated by the TmoAB two-component flevin mononucleotide-dependent monooxygenase system in Acidovorax sp. strain T1[J]. Appl Environ Microbiol, 2017, 83(23): e0l490-e01517. |
| [51] |
Miwa N, Takeda Y, Kuwatsuka S. Plasmid in the degrader of the herbicide thiobencarb (benthiocarb) isolated from soil: a possible mechanism for enrichment of pesticide degraders in soil[J]. J Pestic Sci, 1988, 13(2): 291-293. DOI:10.1584/jpestics.13.291 |
| [52] |
Torra R M, Yajima M, Yamanaka S, et al. Degradation of the herbicides thiobencarb, butachlor and molinate by a newly isolated Aspergillus niger[J]. J Pestic Sci, 2004, 29(3): 214-216. DOI:10.1584/jpestics.29.214 |
| [53] |
Barreiros L, Nogales B, Manaia C M, et al. A novel pathway for mineralization of the thiocarbamate herbicide molinate by a defined bacterial mixed culture[J]. Environ Microbiol, 2003, 5(10): 944-953. DOI:10.1046/j.1462-2920.2003.00492.x |
| [54] |
Duarte M, Ferreiradasilva F, Lünsdorf H, et al. Gulosibacter molinativorax ON4T molinate hydrolase, a novel cobalt-dependent amidohydrolase[J]. J Bacteriol, 2011, 193(20): 5 810-5 816. DOI:10.1128/JB.05054-11 |
| [55] |
Barreiros L, Fernandes A, Ferreira A, et al. New insights into a bacterial metabolic and detoxifying association responsible for the mineralization of the thiocarbamate herbicide molinate[J]. Microbiology, 2008, 154(4): 1 038-1 046. DOI:10.1099/mic.0.2007/015297-0 |
 2018, Vol. 40
2018, Vol. 40


